Toy Batch SLAM Example¶
In this example, we’ll consider a similar example to the one in the toy problems notebook, but this time, we’ll consider it in a batch estimation framework. To increase the complexity of the problem, we will also assume that the landmark positions are unknown, and will be part of the state for estimation. Our goal will be to estimate the robot’s trajectory and the landmark positions using all of the measurements collected. The state to estimate is given by
where similarly to in the previous example \(\mathbf{T}_k \in SE(2)\) denotes the pose of the robot at time \(k\), written as
Additionally, \(\mathbf{p}_a^i \in \mathbb{R}^2\) denotes the position of the \(i\)’th landmark in the inertial frame. Similarly to the previous example, the robot collects wheel odometry measurements, which are used as the input to the process model given by
In this example, the robot will also collects direct measurements of the landmark position, resolved in the body frame of the robot. The measurement model is now a function of one robot pose and one landmark state, such that the measurement of the \(j\)’th landmark at time \(k\) is given by
where \(\mathbf{v}_{jk} \sim \mathcal{N} \left( \mathbf{0}, \mathbf{R}_{jk} \right)\) is white noise. Notice that unlike the previous example, each measurement is a function of two states. To define the measurement model, the CompositeState class can be leveraged. First, we’ll start by defining some parameters that we’ll need for the rest of the example, and then we’ll implement the measurement model.
[1]:
import navlie as nav
import numpy as np
import typing
from pymlg import SO3
### Parameters used for the example
# if true, the information matrix in the batch problem will be inverted to compute the covariance
compute_covariance = True
# If false, will run without noise, and all states initialized to groundtruth
noise = True
# String keys to identify the states
pose_key_str = "x"
landmark_key_str = "l"
# The end time of the simulation
t_end = 20.0
### Defining the measurement model
class PointRelativePositionSLAM(nav.types.MeasurementModel):
def __init__(self, pose_state_id: typing.Any, landmark_state_id: typing.Any, R: np.ndarray):
self.pose_state_id = pose_state_id
self.landmark_state_id = landmark_state_id
self._R = R
def evaluate(self, x: nav.CompositeState):
pose: nav.lib.SE2State = x.get_state_by_id(self.pose_state_id)
landmark: nav.lib.VectorState = x.get_state_by_id(self.landmark_state_id)
r_a = pose.position.reshape((-1, 1))
p_a = landmark.value.reshape((-1, 1))
C_ab = pose.attitude
return C_ab.T @ (p_a - r_a)
def jacobians(self, x: nav.CompositeState):
pose: nav.lib.SE2State = x.get_state_by_id(self._pose_state_id)
landmark: nav.lib.VectorState = x.get_state_by_id(self._landmark_state_id)
r_zw_a = pose.position.reshape((-1, 1))
C_ab = pose.attitude
r_pw_a = landmark.value.reshape((-1, 1))
y = C_ab.T @ (r_pw_a - r_zw_a)
# Compute Jacobian of measurement model with respect to the state
if pose.direction == "right":
pose_jacobian = pose.jacobian_from_blocks(
attitude=-SO3.odot(y), position=-np.identity(r_zw_a.shape[0])
)
elif pose.direction == "left":
pose_jacobian = pose.jacobian_from_blocks(
attitude=-C_ab.T @ SO3.odot(r_pw_a), position=-C_ab.T
)
# Compute jacobian of measurement model with respect to the landmark
landmark_jacobian = pose.attitude.T
# Build full Jacobian
state_ids = [state.state_id for state in x.value]
jac_dict = {}
jac_dict[state_ids[0]] = pose_jacobian
jac_dict[state_ids[1]] = landmark_jacobian
return x.jacobian_from_blocks(jac_dict)
def covariance(self, x: nav.CompositeState):
return self._R
Evaluating The Measurement Model¶
To evaluate this measurement model, we just need to create a CompositeState that contains the robot state and the landmark state, as done below.
[2]:
pose = nav.lib.SE2State(np.array([0.1, 1.0, 2.0]), state_id=pose_key_str)
landmark = nav.lib.VectorState(np.array([1.0, 2.0]), state_id=landmark_key_str)
R = np.identity(2) * 0.01
# Create the measurement model
model = PointRelativePositionSLAM(pose_key_str, landmark_key_str, R)
# Create a composite state
state = nav.lib.CompositeState([pose, landmark])
# Evaluate the model
y = model.evaluate(state)
print(y)
[[ 0.09642014]
[-0.05653507]]
Creating the simulated data¶
Next, we’ll create the simulated data for this example. To use the data generator included in navlie, it is convenient to define the same relative position measurement model, but where the landmarks are included in the state vector. We’ll start by defining some landmarks and creating the relative position measurement model with known landmarks. Then, we’ll create the process model using the BodyFrameVelocity process model defined in navlie.
[3]:
class PointRelativePosition(nav.types.MeasurementModel):
def __init__(self, landmark_position: np.ndarray, landmark_id: int, R: np.ndarray):
self.landmark_position = landmark_position.reshape((-1, 1))
self.landmark_id = landmark_id
self._R = R
def evaluate(self, x: nav.lib.SE2State):
r_a = x.position.reshape((-1, 1))
p_a = self.landmark_position
C_ab = x.attitude
return C_ab.T @ (p_a - r_a)
def covariance(self, x: nav.CompositeState):
return self._R
# Now, create some landmarks arranged in a circle and create a list of
# measurement models, one for each landmark
landmark_positions = [np.array([3.0 * np.cos(theta), 3.0 * np.sin(theta)]) for theta in np.linspace(0, 2*np.pi, 10)]
landmarks = [nav.lib.VectorState(landmark, state_id=f"{landmark_key_str}{i}") for i, landmark in enumerate(landmark_positions)]
R = np.identity(2) * 0.1
meas_models = [PointRelativePosition(l.value, l.state_id, R) for l in landmarks]
# Create the process model
Q = np.identity(3) * 0.4
process_model = nav.lib.BodyFrameVelocity(Q)
# Input profile
input_profile = lambda t, x: np.array(
[np.cos(0.1 * t), 1.0, 0]
)
# Generate the data
x0 = nav.lib.SE2State(np.array([0, 0, 0]))
dg = nav.DataGenerator(
process_model,
input_profile,
Q,
input_freq=100,
meas_model_list=meas_models,
meas_freq_list=[10] * len(meas_models),
)
gt_poses, input_list, meas_list = dg.generate(x0, start=0.0, stop=t_end, noise=noise)
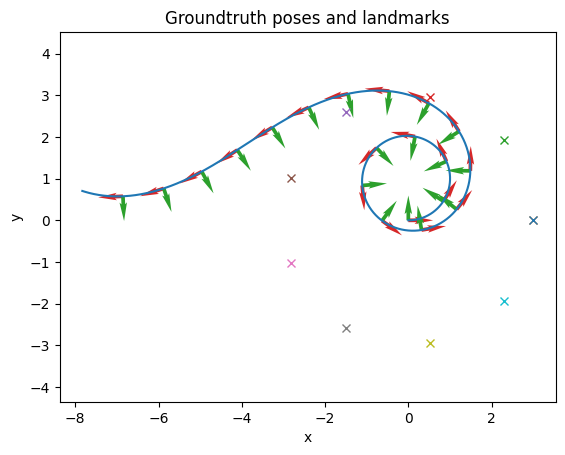
# Plot the true state
import matplotlib.pyplot as plt
fig, ax = nav.plot_poses(gt_poses, step=100)
for landmark in landmarks:
ax.plot(landmark.value[0], landmark.value[1], 'x')
ax.set_title("Groundtruth poses and landmarks")
ax.set_xlabel("x")
ax.set_ylabel("y")
plt.show()
Batch Estimation: From Weighted Nonlinear Least Squares to Unweighted Nonlinear Least Squares¶
Now, we wish to estimate the full state of the system using all of the inputs and all of the measaurements. To do this, we want to define a nonlinear least squares problem of the form
where \(N\) is the number of error terms in the problem, and each \(\mathbf{e}_i (\mathcal{X})\) is an error term that is function of the state. Additionally, \(\mathbf{W}_i\) is a weight matrix, generally the inverse covariance of the error. navlie has a nonlinear least squares solver Problem that can be used to solve general nonlinear least squares problems. However, the solver can only handle unweighted nonlinear least squares problems of the form
Thankfully, converting the weighted nonlinear least squares problem to an equivalent unweighted problem is possible. Consider the Cholesky factorization of \(\mathbf{W}\), given by
Defining \(\tilde{\mathbf{e}_i} = \mathbf{L}_i \mathbf{e}_i\), we can rewrite the original weighted cost function as
Thus, for each error term in the problem, the user must pay careful attention to weight the error by the square root of the inverse covariance matrix of that particular error term.
Now that we’ve seen how to convert the original weighted nonlinear least squares problem into a nonweighted one, let’s explore how to define nonlinear least squares problems in navlie.
Extracting the Estimates and the Covariances¶
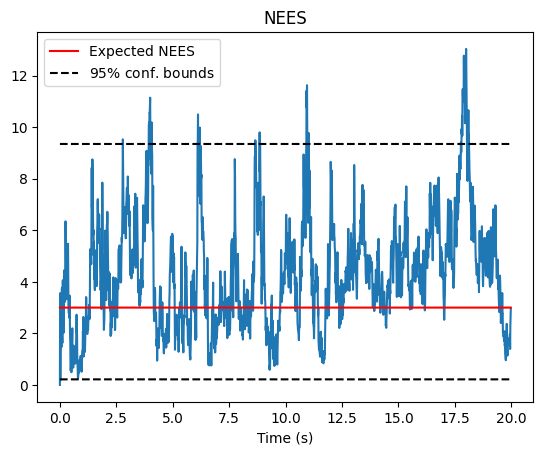
To extract the covariance, we can use thee problem.get_covariance_block() method, which will extract the marginal covariance of a particular variable based on the state ID of that variable. Note that this is useful for visualizing three sigma bounds and individual NEES values for subsets of the state, but sometimes it is also useful to compute the NEES for the entire trajectory. The problem class also outputs the full information matrix of the problem, and the user can choose to manipulate
this themselves after the optimization has completed.
[9]:
# Extract estimates
poses_results_list: typing.List[nav.types.StateWithCovariance] = []
for pose in init_pose_est:
state = variables_opt[pose.state_id]
if compute_covariance:
# Extract the covariance for only this current pose state
cov = problem.get_covariance_block(pose.state_id, pose.state_id)
else:
cov = np.identity(3)
poses_results_list.append(nav.types.StateWithCovariance(state, cov))
landmarks_results_list: typing.List[nav.types.StateWithCovariance] = []
for landmark in init_landmark_est:
state = variables_opt[landmark.state_id]
if compute_covariance:
cov = problem.get_covariance_block(landmark.state_id, landmark.state_id)
else:
cov = np.identity(2)
landmarks_results_list.append(nav.types.StateWithCovariance(state, cov))
# Postprocess the results and plot
gaussian_result_list = nav.GaussianResultList(
[nav.GaussianResult(poses_results_list[i], gt_poses[i]) for i in range(len(poses_results_list))],
)
# Plot NEES
fig, axs = nav.plot_nees(gaussian_result_list)
axs.set_xlabel("Time (s)")
axs.set_title("NEES")
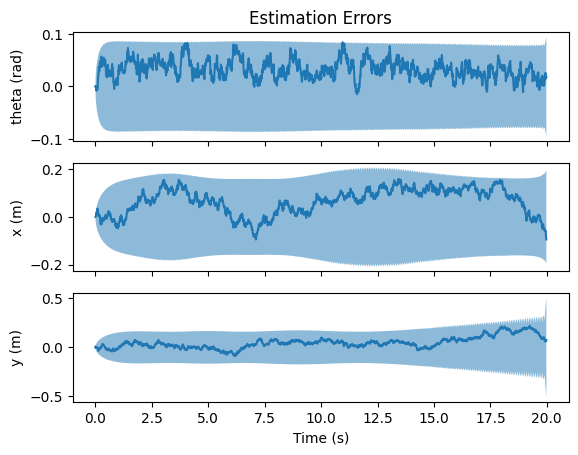
fig, axs = nav.plot_error(gaussian_result_list)
axs[0].set_title("Estimation Errors")
axs[0].set_ylabel("theta (rad)")
axs[1].set_ylabel("x (m)")
axs[2].set_ylabel("y (m)")
axs[2].set_xlabel("Time (s)")
plt.show()
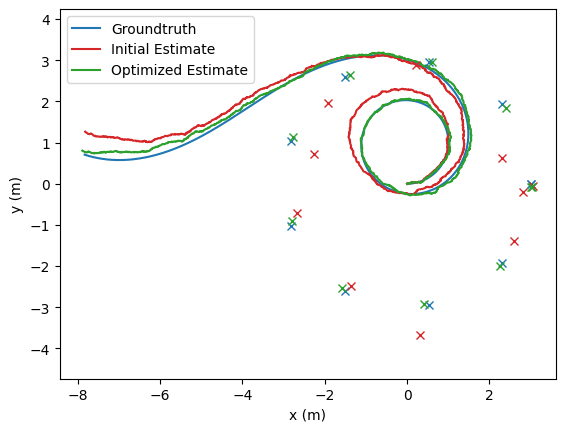
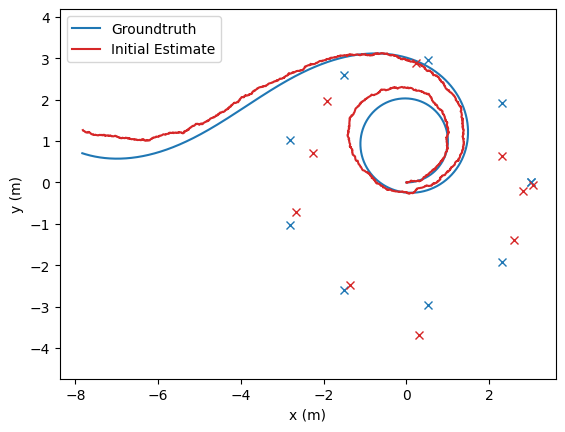
# Plot the initial estimate, optimized estimates, and groundtruth
opt_poses: typing.List[nav.lib.SE2State] = [state.state for state in poses_results_list]
fig, ax = nav.plot_poses(gt_poses, step = None, line_color='tab:blue', label="Groundtruth")
fig, ax = nav.plot_poses(init_pose_est, step=None, ax=ax, line_color='tab:red', label="Initial Estimate")
fig, ax = nav.plot_poses(opt_poses, step=None, ax=ax, line_color='tab:green', label="Optimized Estimate")
opt_landmarks: typing.List[nav.lib.VectorState] = [state.state for state in landmarks_results_list]
for landmark in landmarks:
ax.plot(landmark.value[0], landmark.value[1], 'tab:blue', marker='x')
for landmark in init_landmark_est:
ax.plot(landmark.value[0], landmark.value[1], 'tab:red', marker='x')
for landmark in opt_landmarks:
ax.plot(landmark.value[0], landmark.value[1], 'tab:green', marker='x')
ax.set_xlabel("x (m)")
ax.set_ylabel("y (m)")
ax.legend()
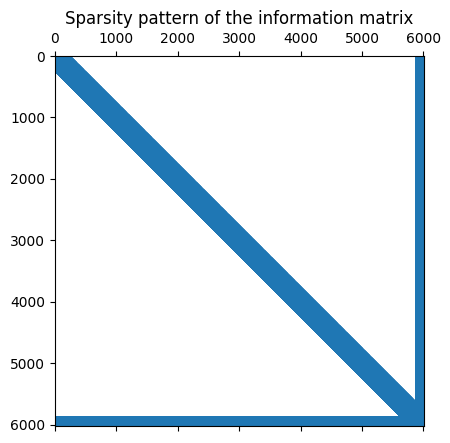
# Visualize the sparsity pattern of the information matrix
fig, ax = plt.subplots()
ax.set_title("Sparsity pattern of the information matrix")
ax.spy(opt_results["info_matrix"])
plt.show()